Functional Bioinformatics Algorithms: Part 2
September 23, 2020 3 Comments
Pressing on with more bioinformatic algorithms implemented in a functional style, the next algorithm found in Bioinformatics Algorithms by Compeau and Pevzner is to find the most frequent pattern in a string of text.
I started writing in the imperative style from the book like so (the length of the substring to be found is called the “k-mer” so it gets the parameter name “k”)
type Count = {Text:string; PatternCount:int}
let frequentWords (text:string) (k:int) =
let patternCount (text:string) (pattern:string) =
text
|> Seq.windowed pattern.Length
|> Seq.map(fun c -> new string(c))
|> Seq.filter(fun s -> s = pattern)
|> Seq.length
let counts = new List<Count>()
for i = 0 to text.Length-k do
let pattern = text.Substring(i,k)
let patternCount = patternCount text pattern
let count = {Text=text; PatternCount=patternCount}
counts.add(count)
counts |> Seq.orderByDesc(fun c -> c.PatternCount)
let maxCount = counts|> Seq.head
let frequentPatterns = new List<Count>()
for i = 0 to counts.length
if count.[i].PatternCount = maxCount then
frequentPatterns.add(count.[i])
else
()But I gave up because, well, the code is ridiculous. I went back to the original pattern count algorithms written in F# and then added in a block to find the most frequent patterns:
let frequentWords (text:string) (k:int) =
let patternCounts =
text
|> Seq.windowed k
|> Seq.map(fun c -> new string(c))
|> Seq.countBy(fun s -> s)
|> Seq.sortByDescending(fun (s,c) -> c)
let maxCount = patternCounts |> Seq.head |> snd
patternCounts
|> Seq.filter(fun (s,c) -> c = maxCount)
|> Seq.map(fun (s,c) -> s)The VS Code linter was not happy with my Seq.countBy implementation… but it works. I think the code is explanatory:
- window the string for the length of k
2. do a countyBy on the resulting substrings
3. sort it by descending, find the top substring amount
4. filter the substring list by that top substring count.
The last map returns just the pattern and leaves off the frequency, which I think is a mistake but is how the book implements it. Here is an example of the frequentWords function in action:
let getRandomNuclotide () =
let dictionary = ["A";"C";"G";"T"]
let random = new Random()
dictionary.[random.Next(4)]
let getRandomSequence (length:int) =
let nuclotides = [ for i in 0 .. length -> getRandomNuclotide() ]
String.Join("", nuclotides)
let largerText = getRandomSequence 1000000
let currentFrequentWords = frequentWords largerText 9
currentFrequentWords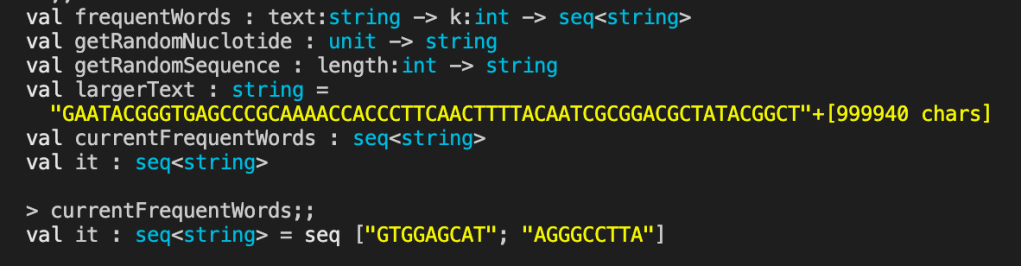
I didn’t set the seed value for generating the largerText string so the results will be different each time.
Gist is here
Pingback: Dew Drop – September 24, 2020 (#3282) | Morning Dew
Pingback: F# Weekly #39, 2020 – Giraffe is 50-100% faster of .NET 5 & F# eXchange 2020 – Sergey Tihon's Blog
> The VS Code linter was not happy with my Seq.countBy implementation
This is because (fun s -> s) is a function called “id”, so instead of Seq.countBy (fun s -> s), try Seq.countBy id.